Test Note Again
Let’s try syndication again. This account will likely have a lot of test posts before I get things making sense.
Let’s try syndication again. This account will likely have a lot of test posts before I get things making sense.
I’m testing syndication here! Hopefully this posts to my site and Mastodon and allows for webmentions back to my site.
We developed an atlas for adaptive smoothing in the brain based on white matter tracts.
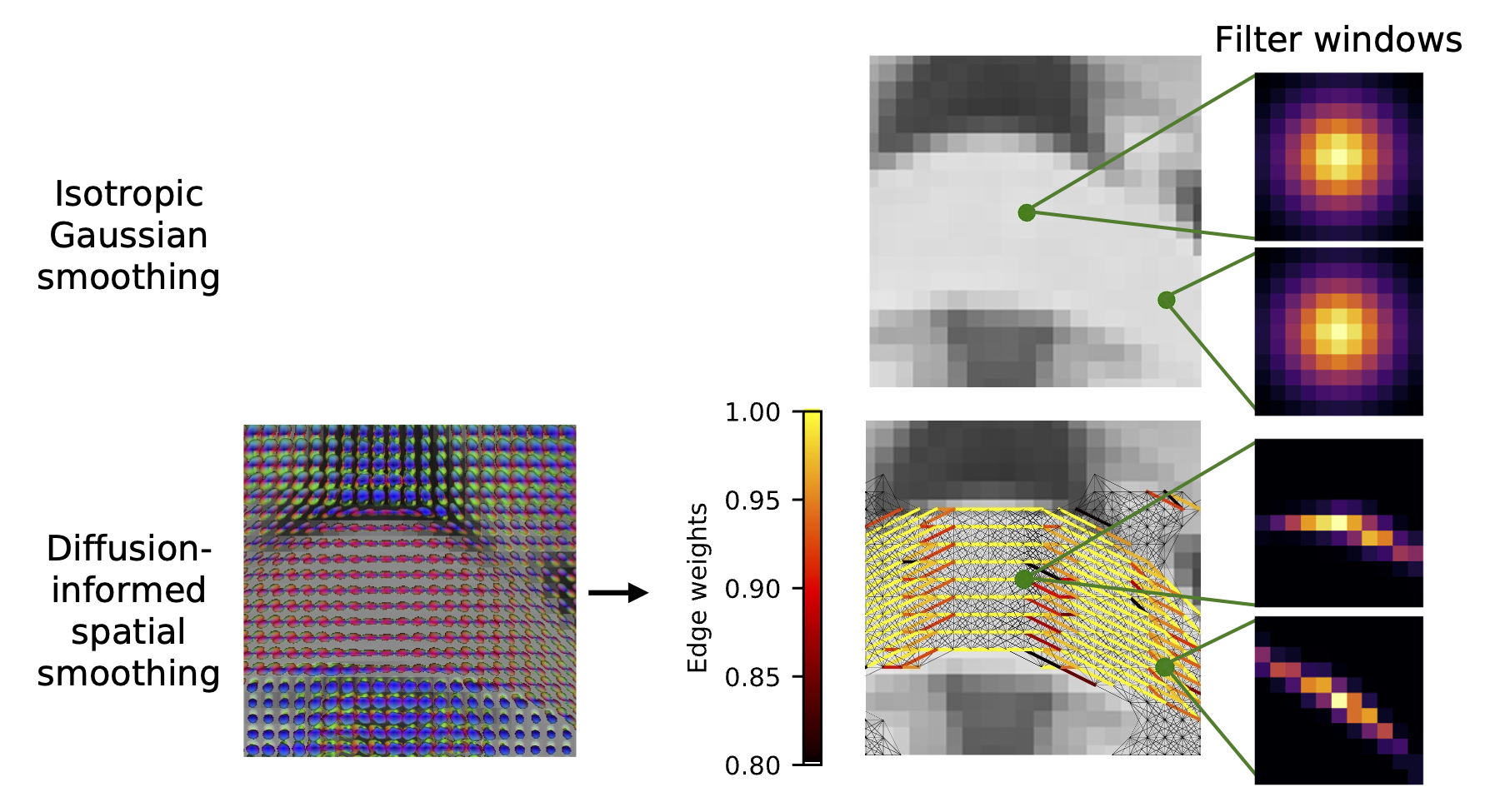 Using our atlas, we can adaptively smooth functional MRI data based on white matter tracts, which better preserves the underlying signal present compared to traditional smoothing methods.
Using our atlas, we can adaptively smooth functional MRI data based on white matter tracts, which better preserves the underlying signal present compared to traditional smoothing methods.
Adam M. Saunders, Gaurav Rudravaram, Nancy R. Newlin, Michael E. Kim, John C. Gore, Bennett A. Landman, and Yurui Gao. A 4D atlas of diffusion-informed spatial smoothing windows for BOLD signal in white maxtter. SPIE Medical Imaging: Image Processing, February 2025. [doi:10.1117/12.3047240].
Typical methods for preprocessing functional magnetic resonance images (fMRI) involve applying isotropic Gaussian smoothing windows to denoise blood oxygenation level-dependent (BOLD) signals, a process which spatially smooths white matter signals that occur along anisotropically-oriented fibers. Abramian et al. have proposed diffusion-informed spatial smoothing (DSS) filters to smooth white matter in a physiologically-informed manner. However, these filters rely on paired diffusion MRI and fMRI data, which are not always available. Here, we create DSS windows for smoothing fMRI data in the white matter based on the Human Connectome Project Young Adult population-averaged atlas of fiber orientation distribution functions. We smooth fMRI data from 63 subjects using the atlas-based DSS windows and compare the results with fMRI data smoothed with isotropic Gaussian windows at 1.04 mm full-width half-max (FWHM) and 3 mm FWHM. Compared to isotropic Gaussian windows, the atlas-based DSS windows result in fMRI data with a significantly higher local functional connectivity measured with regional homogeneity (ReHo, p < 0.001). The DSS atlas results in biologically informed regions of interest identified through independent component analysis that more closely agree with regions from a diffusion MRI-based white matter atlas. The DSS atlas generated here allows for diffusion-informed smoothing of fMRI data when additional diffusion MRI data are not available. The DSS atlas and code are available online (https://github.com/MASILab/dss_fmri_atlas).
We developed a method for adaptive spatial smoothing based on the vasculature in the brain.
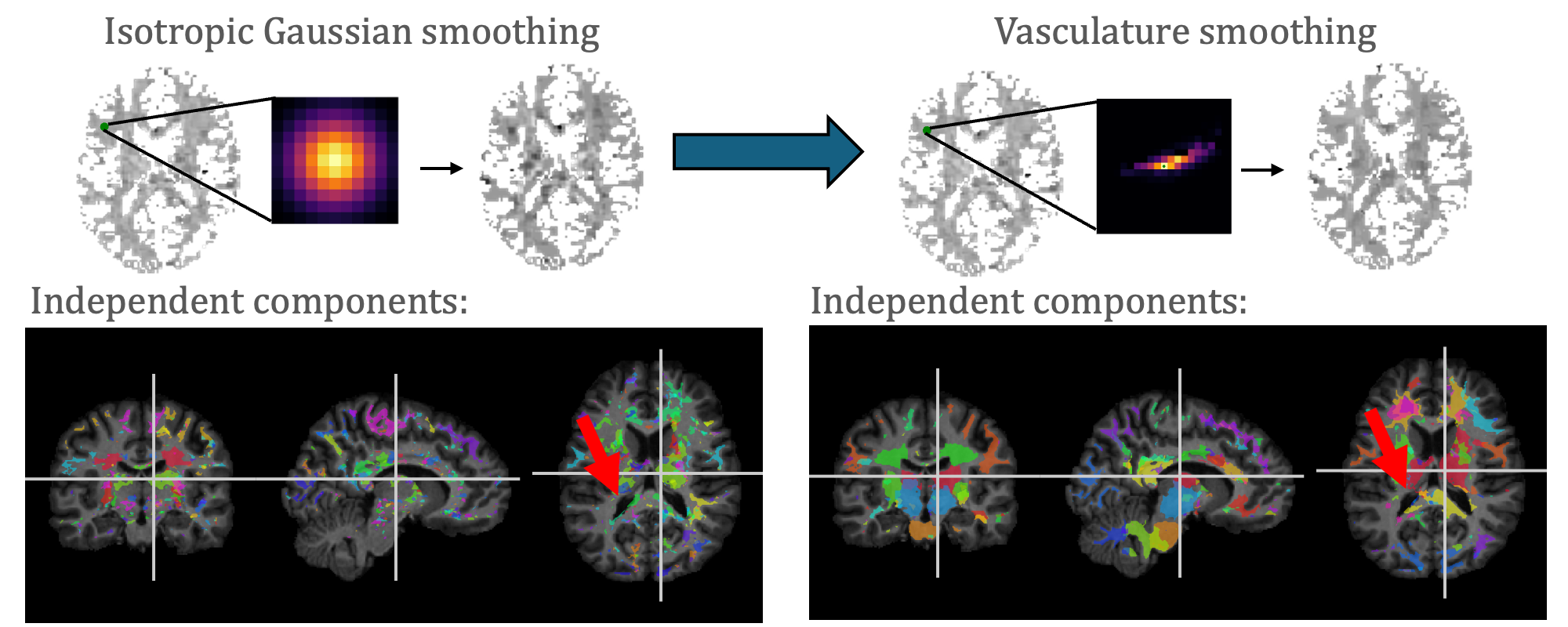 With our adaptive vasculature smoothing, we are able to identify more anatomically-informed independent components from fMRI data.
With our adaptive vasculature smoothing, we are able to identify more anatomically-informed independent components from fMRI data.
Adam M. Saunders, Michael E. Kim, Kurt G. Schilling, John C. Gore, Bennett Papers A. Landman, and Yurui Gao. Vasculature-informed spatial smoothing of white matter functional magnetic resonance imaging. SPIE Medical Imaging: Image Processing, February 2025. [doi: 10.1117/12.3047240]. Robert F. Wagner All-Conference Best Student Paper Finalist.
Blood oxygenation level-dependent (BOLD) signals in white matter in the brain are anisotropically oriented, so that typical isotropic Gaussian spatial smoothing (GSS) of functional magnetic resonance images (fMRI) blurs across anatomical distributions. Abramian et al. developed a graph signal processing approach to smooth fMRI data along white matter fibers using diffusion MRI (diffusion-informed spatial smoothing, DSS). BOLD signals are modulated by the volume and oxygenation of blood carried by the vasculature, so we extend this method to provide vasculature-informed spatial smoothing (VSS). We collected susceptibility-weighted images and applied a Frangi filter to identify the peak vasculature direction in each voxel, alongside co-registered diffusion MRI and resting-state fMRI, weighting the VSS graph by the agreement of the vasculature directions aligned onto the graph’s edges. We acquired resting-state fMRI at 7T using a repetition time of 1.5 seconds and 400 time points. Applying the DSS and VSS filters significantly increased the local functional connectivity measured using regional homogeneity (ReHo) compared to GSS (𝑝 < 0.01 using a paired t-test), but not when comparing DSS and VSS (p = 0.06). Independent component analysis resulted in less noisy components that agree better with labels from a white matter atlas with a significantly higher Dice score from the VSS filter compared to GSS (p < 0.05 using the Mann-Whitney U-test), and the VSS filter and DSS filter performed comparably (p = 0.06). In this pilot analysis, we find that fMRI data smoothed using VSS are comparable to results generated using DSS. The filtering code is available online (https://github.com/MASILab/vss_fmri).
We found and corrected a bias in quantitative T1 methods using a patch-based deep learning model.
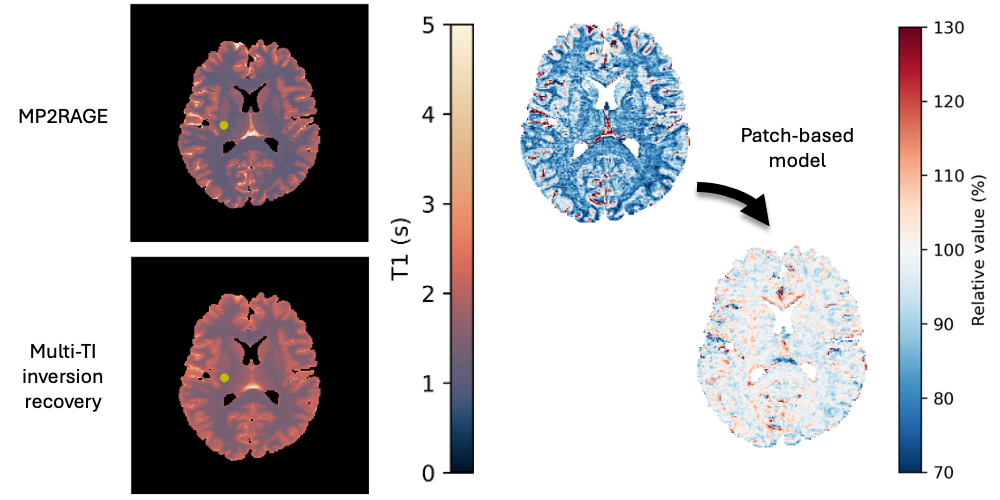 We found a bias between two methods for mapping quantitative values of T1 from MRI, and we corrected that bias with a patch-based deep learning model.
We found a bias between two methods for mapping quantitative values of T1 from MRI, and we corrected that bias with a patch-based deep learning model.
Adam M. Saunders, Michael E. Kim, Chenyu Gao, Lucas W. Remedios, Aravind R. Krishnan, Kurt G. Schilling, Kristin P. O’Grady, Seth A. Smith, and Bennett A. Landman. Comparison and calibration of MP2RAGE quantitative T1 values to multi-TI inversion recovery T1 values. Magnetic Resonance Imaging, 117, 110322 (2025). [doi:10.1016/j.mri.2025.110322].
While typical qualitative T1-weighted magnetic resonance images reflect scanner and protocol differences, quantitative T1 mapping aims to measure T1 independent of these effects. Changes in T1 in the brain reflect structural changes in brain tissue. Magnetization-prepared two rapid acquisition gradient echo (MP2RAGE) is an acquisition protocol that allows for efficient T1 mapping with a much lower scan time per slab compared to multi-TI inversion recovery (IR) protocols. We collect and register B1-corrected MP2RAGE acquisitions with an additional inversion time (MP3RAGE) alongside multi-TI selective inversion recovery acquisitions for four subjects. We use a maximum a posteriori (MAP) T1 estimation method for both MP2RAGE and compare to typical point estimate MP2RAGE T1 mapping, finding no bias from MAP MP2RAGE but a sensitivity to B1 inhomogeneities with MAP MP3RAGE. We demonstrate a tissue-dependent bias between MAP MP2RAGE T1 estimates and the multi-TI inversion recovery T1 values. To correct this bias, we train a patch-based ResNet-18 to calibrate the MAP MP2RAGE T1 estimates to the multi-TI IR T1 values. Across four folds, our network reduces the RMSE significantly (white matter: from 0.30 +/- 0.01 seconds to 0.11 +/- 0.02 seconds, subcortical gray matter: from 0.26 +/- 0.02 seconds to 0.10 +/- 0.02 seconds, cortical gray matter: from 0.36 +/- 0.02 seconds to 0.17 +/- 0.03 seconds). Using limited paired training data from both sequences, we can reduce the error between quantitative imaging methods and calibrate to one of the protocols with a neural network.
We published our work on creating eye atlases from low-resolution MRI.
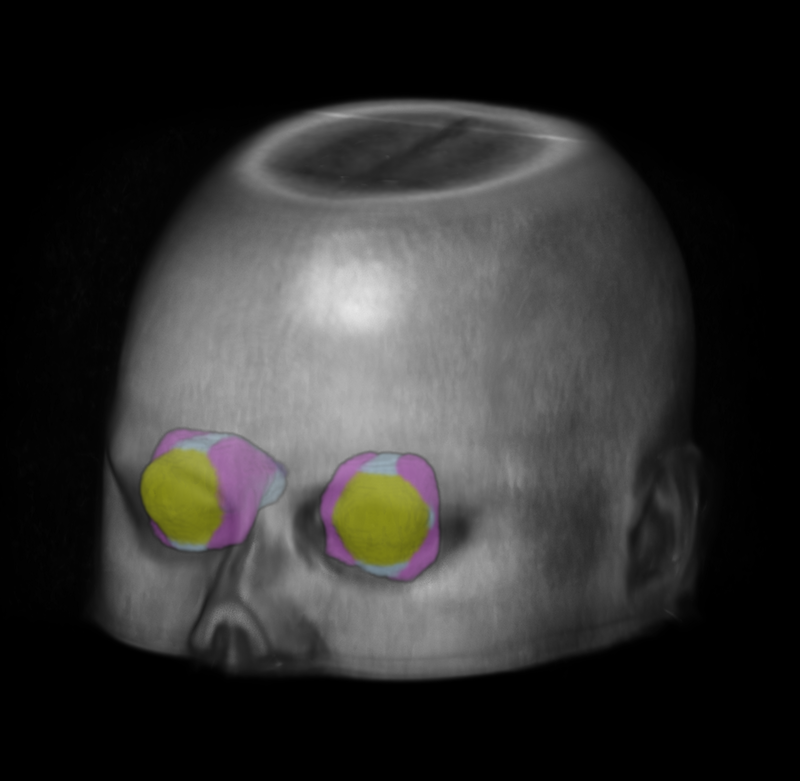 We created standardized reference images for the eye using low-resolution MRI.
We created standardized reference images for the eye using low-resolution MRI.
Ho Hin Lee*, Adam M. Saunders*, Michael E. Kim, Samuel W. Remedios, Lucas W. Remedios, Yucheng Tang, Qi Yang, Xin Yu, Shunxing Bao, Chloe Cho, Louise A. Mawn, Tonia S. Rex, Kevin L. Schey, Blake E. Dewey, Jeffrey M. Spraggins, Jerry L. Prince, Yuankai Huo, Bennett A. Landman, Super-resolution multi-contrast unbiased eye atlases with deep probabilistic refinement. Journal of Medical Imaging, 11(6), 064004 (2024). *Equal contribution. [doi:10.1117/1.JMI.11.6.064004]
Purpose: Eye morphology varies significantly across the population, especially for the orbit and optic nerve. These variations limit the feasibility and robustness of generalizing population-wise features of eye organs to an unbiased spatial reference. Approach: To tackle these limitations, we propose a process for creating high-resolution unbiased eye atlases. First, to restore spatial details from scans with a low through-plane resolution compared with a high in-plane resolution, we apply a deep learning-based super-resolution algorithm. Then, we generate an initial unbiased reference with an iterative metric-based registration using a small portion of subject scans. We register the remaining scans to this template and refine the template using an unsupervised deep probabilistic approach that generates a more expansive deformation field to enhance the organ boundary alignment. We demonstrate this framework using magnetic resonance images across four different tissue contrasts, generating four atlases in separate spatial alignments. Results: When refining the template with sufficient subjects, we find a significant improvement using the Wilcoxon signed-rank test in the average Dice score across four labeled regions compared with a standard registration framework consisting of rigid, affine, and deformable transformations. These results highlight the effective alignment of eye organs and boundaries using our proposed process. Conclusions: By combining super-resolution preprocessing and deep probabilistic models, we address the challenge of generating an eye atlas to serve as a standardized reference across a largely variable population.
I presented a TED-style talk highlighting the aims of my honors thesis research into diabetic retinopathy imaging and deep learning.
 Deep Learning for a Healthier World: Detecting and Grading Diabetic Retinopathy
Deep Learning for a Healthier World: Detecting and Grading Diabetic Retinopathy
In late April 2023, I presented a talk at the first-ever Honors Thesis Signature Talks at Stander Symposium at the University of Dayton. These talks featured four undergraduate honors researchers presenting their work in a TED-style format. In my talk, Deep Learning for a Healthier World: Detecting and Grading Diabetic Retinopathy, I discuss my family’s experience with diabetic retinopathy, and how it inspired me to pursue medical imaging research. I explore how we can use deep learning models to learn patterns from medical images, making them a useful tool for detecting diseases like diabetic retinopathy. We can apply innovative transformations that recolor, resize, crop and transform the images to allow the deep learning model to learn to detect diseases more easily.
In addition, my honors thesis paper is available online as well: Methods for Exploiting High-Resolution Imagery for Deep Learning-Based Diabetic Retinopathy Detection and Grading
Abstract
Diabetic retinopathy is a disease that affects the eyes of people with diabetes, and it can cause blindness. To diagnose diabetic retinopathy, ophthalmologists image the back surface of the inside of the eye, a process referred to as fundus photography. Ophthalmologists must then diagnose and grade the severity of diabetic retinopathy by analyzing details in the image, which can be difficult and time-consuming. Alternatively, due to the availability of labeled datasets containing fundus images with diabetic retinopathy, AI methods like deep learning can provide automated detection and grading algorithms. We show that the resolution of an image has a large effect on the accuracy of grading algorithms. So, we study several techniques to increase the accuracy of the algorithm by taking advantage of higher-resolution data, including using a region of interest as the input and applying an image transformation to make the circular fundus image square. While none of our proposed methods result in an increase in performance for grading diabetic retinopathy, the circle to square transformation results in an increase in accuracy and AUC for detection of diabetic retinopathy. This work provides a useful starting point for future research aimed at increasing the resolution content in a fundus image.
I graduated summa cum laude from the University of Dayton with a Bachelor of Electrical Engineering.
 I celebrated my graduation from the University of Dayton
I celebrated my graduation from the University of Dayton
I’m proud to announce I have officially graduated from the University of Dayton! I have graduated with a Bachelor of Electrical Engineering with a minor in Mathematics. It’s been a great few years, and I’m excited for what’s next.
I graduated summa cum laude. In addition, I was the recipient of the Thomas R. Armstrong Award of Excellence for Outstanding Electrical Engineering Achievement. Along with my twin brother Nick, I also received the Department of Music’s Senior Award for Outstanding Collaborative Pianist. I was very honored to receive these awards, and I think they highlight one of the main reasons I first chose to attend UD - I worked hard to study engineering while also continuing to include music in my life. I hope to be able to continue incorporating music into my life, especially as a piano accompanist.
The next step for me is moving to Nashville to begin a PhD program at Vanderbilt University! I’m super excited to continue performing medical imaging research. I’m overwhelmed with gratitude for everyone who has helped get me to this point. The journey to becoming an electrical engineer has not been easy, but it has certainly been worth it!
I accepted a PhD offer from Vanderbilt University at the MASI Lab under Dr. Bennett Landman.
I’m excited to share that I have accepted a PhD offer from Vanderbilt University! I will be joining the Medical-image Analysis and Statistical Interpretation (MASI) Lab under Dr. Bennett Landman.
This lab is a part of Vanderbilt Institute for Surgery and Engineering (VISE), an exciting, cross-disciplinary set of labs that works in the intersection between medicine, computer science, engineering, and more. So, I’ll officially be a part of the newly-formed Department of Electrical and Computer Engineering, and I will also have the chance to work alongside people across disciplinary boundaries.
I’m ready to move to Nashville and continuing pursuing my goal of earning a PhD!
 I visited Centennial Park in Nashville while on a visit to Vanderbilt
I visited Centennial Park in Nashville while on a visit to Vanderbilt
I attended SPIE Medical Imaging for the first time and presented research I performed last summer on comparing whole-slide image classification algorithms.
 I presented at SPIE Medical Imaging for the first time
I presented at SPIE Medical Imaging for the first time
I recently got the chance to travel to San Diego to present research I performed at Oak Ridge National Laboratory at the 2023 SPIE Medical Imaging conference. This was the first professional conference I’ve attended, and getting the chance to go as an undergraduate was a great experience. I was lucky enough to receive a student travel grant from SPIE to attend.
I was definitely a bit nervous at first to be attending the conference by myself. In fact, I had never traveled alone. However, everyone at the conference was very friendly. I got to meet lots of people from all over the world - Italy, the UK, the Netherlands, and all over the US as well! It was so refreshing to meet people working in medical imaging, especially other students. A highlight for me was talking to students from the MASI Lab at Vanderbilt University, as I’ve applied to join this lab next year as a PhD student.
As someone just starting out in medical imaging, it was very nice to see all of the interesting research going on. The keynote speakers were really fantastic. I particularly enjoyed hearing Zachary Lipton from Carnegie Mellon talk about how the attention mechanism is not a great interpretability measure for deep learning models. If I’m summarizing his talk correctly, his team has found that we can have vastly different attention weights and still get the same accuracy. This result was a bit sombering for people like me who have advertized the attention mechanism as a way of boosting interpretability for image classification. Overall, the talks were very thought-provoking and introduced me to a lot of new ideas.
I got to present the work I performed at Oak Ridge National Laboratory titled “A comparison of histopathology imaging comprehension algorithms based on multiple instance learning.” In this work, we used a supercomputer to compare several multiple instance learning algorithms for whole-slide image classification of cancer datasets. We found that algorithms using the attention mechanism all performed better than those without. It was great to be able to present my work to people who really understood the problem. I had some great conversations with people about what this work meant and how it compared with other researchers’ findings.
I also got to explore San Diego a bit. I went to Mission Beach with a group of students and got to see the Pacific Ocean for the first time! It was cold and rainy - very unseasonable weather for San Diego. I also got to go to Old Town and do some shopping and eating, which was nice.
SPIE Medical Imaging was a great conference, and they were a particularly warm and welcoming group for a first-timer. I hope I’m able to return sometime in the future!
I spent the summer at Oak Ridge National Lab, performing exciting research into designing machine learning algorithms for cancer detection from whole-slide i...

How many people can say they got the chance to use one of the fastest computers in the world to detect cancer? I was extremely lucky to be able to do just that this summer at Oak Ridge National Laboratory in Tennessee. I participated in the Department of Energy’s Science Undergraduate Laboratory Internship (SULI) program, and the experience was incredible.
This past June, I packed up my things and headed down to Knoxville, Tennessee, where I would spend the next few months performing research at nearby Oak Ridge National Laboratory (ORNL). ORNL is one of the Department of Energy’s largest national labs, with nearly 6,000 staff that span a wide variety of research interests from nuclear physics to soil science. ORNL has a fascinating history as well — it was one of the main research sites that powered the Manhattan Project.
The SULI program allows students to spend 10 weeks performing research alongside a mentor at the lab. I worked under Dr. Hong-Jun Yoon to compare machine learning algorithms for analyzing whole-slide images, which are large digital scans of tissue samples. I used Summit, the second-fastest supercomputer in the United States and fourth-fastest in the world, to train and test machine learning models on a massive scale. We successfully detected and subtyped cancers from several datasets.
Along with gaining technical skills in medical image processing and high-performance computing, I also learned how to communicate my results to the wider scientific community. We submitted a manuscript to the 2023 SPIE Medical Imaging conference, and it hopefully will be my first publication. Also, at the ORNL Summer Intern Symposium, my poster won Best Poster in the Computing and Computational Sciences Directorate! You can see my poster and read more about the research I did this summer here.
Outside of my research, I toured several of the unique facilities on campus. I visited X-10, the world’s first continuously operating nuclear reactor. Seeing the name “Oppenheimer” on one of the nameplates was sombering — I was in the former offices of some of the world’s greatest physicists! I also got to see Frontier, the fastest (and greenest) supercomputer in the world!
 The graphite reactor at ORNL is the world’s first continuously operating nuclear reactor
The graphite reactor at ORNL is the world’s first continuously operating nuclear reactor
Getting to spend the summer at ORNL was invigorating, and I developed a passion for medical image processing that I want to pursue further. I consider myself very fortunate for being able to participate in the SULI program, and I hope that I will be able to return to ORNL sometime in the future!